Coffee is one of daily drink all over the world. Coffea arabica, which is the most popular species for current coffee plantation has a natural hybrid genomes (4n=44) originated from Coffea canephora and Coffea eugenioides. There is only one natural hybrid species in Coffea genus. Because of it, its genome cannot be deciphered easily using current recent biotechnologies. So international consortium of coffee genome selected C. canephora genome for understanding coffee genome more in detail; however, C. arabica and C. canpehora have already been fully evolved into the independent species (See table 1) so that we still need genome sequences of C. arabica to understand about coffee genome correctly.
Table 1. Number of variations between C. canephora and C. arabica genomes
| - | C. arabica subsp. typica | C. arabica subsp. kona |
| Mapping ratio | 68.31% | 66.90% |
| SNPs | 4,833,513 | 4,207,675 |
| Insertions | 253,464 | 207,902 |
| Deletions | 240,574 | 199,377 |
As a first step, we sequenced C. arabica subsp. typica genome as a first reference genome. Currently we get genome assembly version 0.2 presenting that total length of genome is 1.02Gbp and N50 is 2,586 bp without mate-pair sequences.
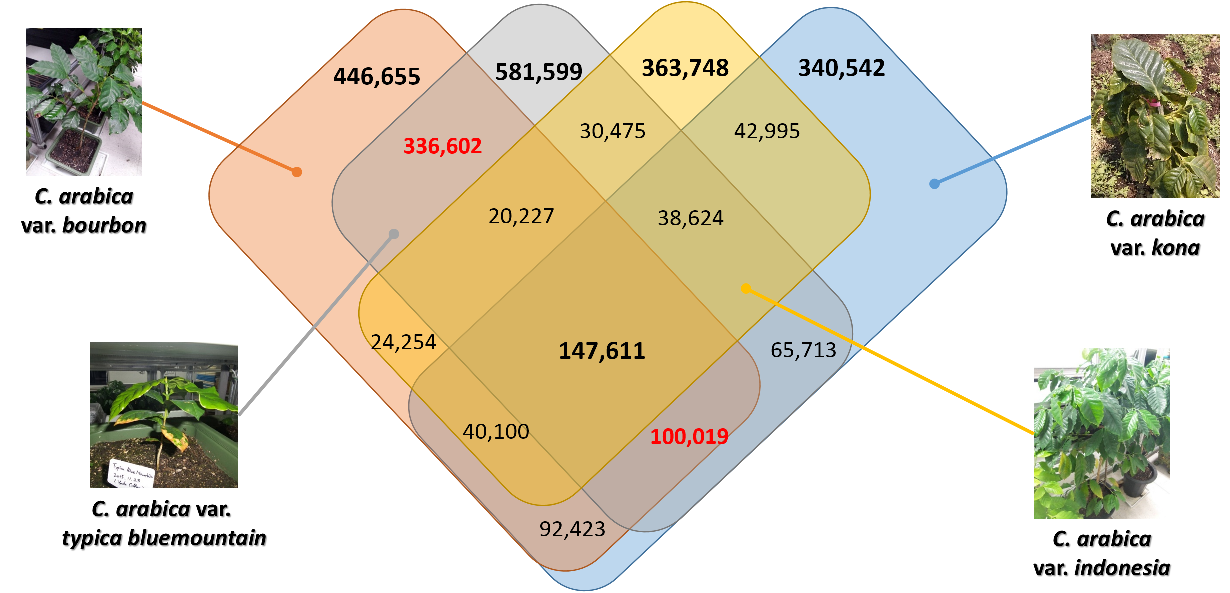
Due to large scale of coffee plantation, there are many subspecies (or variaties or breeding strains) of C. arabica. One of major purpose of this genome project is understanding characteristics of coffee genomes at subspecies level. To archive it, we sequenced five different C. arabica subspecies and compared them with C. arabica subsp. typica genome.
Figure 1. Number of variations (SNPs and INDELs) among five subspecies of C. arabica. Reference genome is C. arabica subsp. typica